Through this portal, you can explore and interactively visualise ProteoCast predictions for Drosophila melanogaster proteome, including all protein isoforms (proteoforms) resulting from alternative splicing.
Try it out with your favorite protein! Please provide FlyBase protein ID (FBpp) or proteoform symbol (i.e. yki-PF) in the search dialog below.
For more information and resources, please refer to the documentation.
How it works:
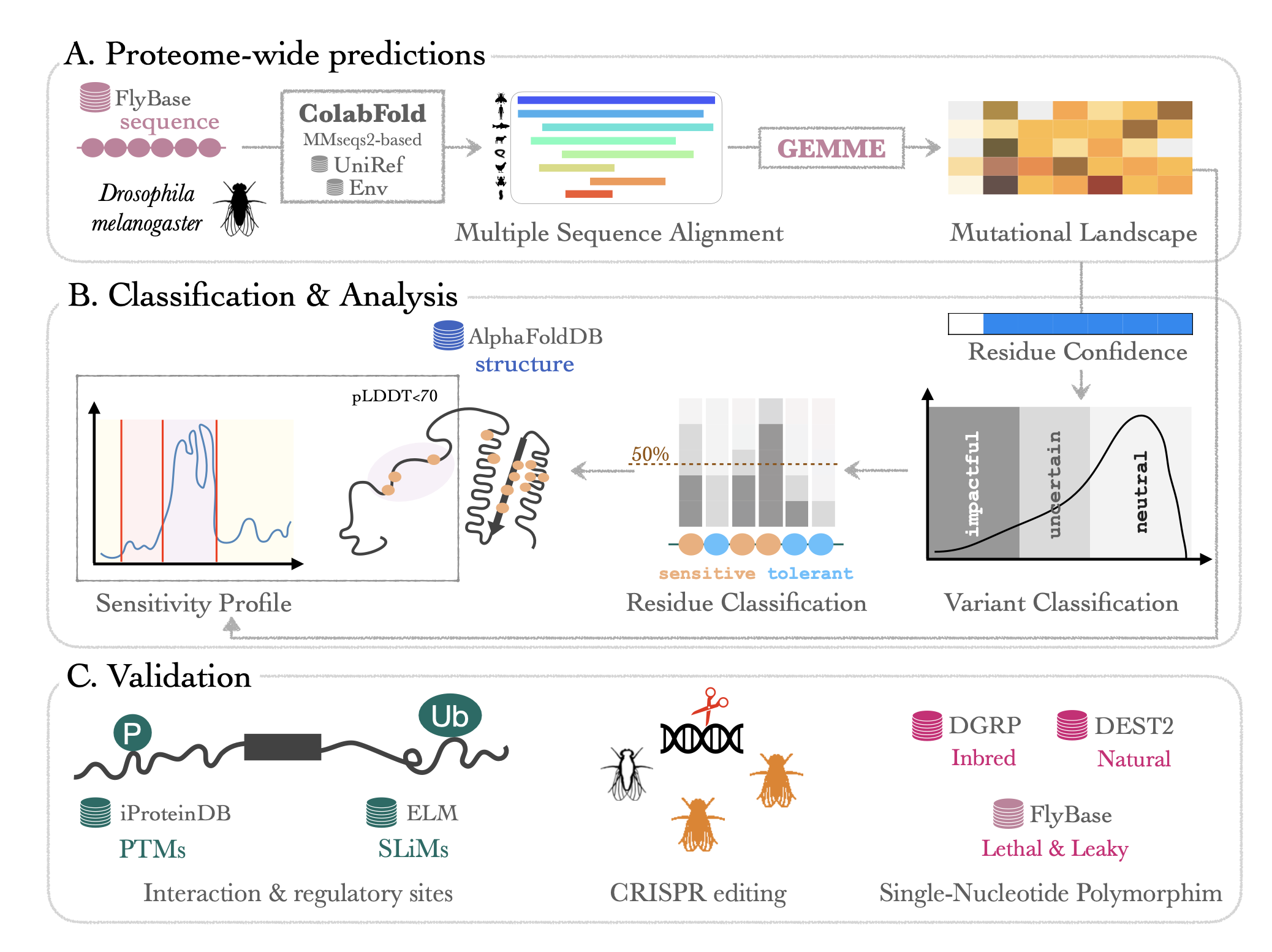
- ProteoCast performed proteome-wide predictions of all possible amino acid variants using GEMME (Laine et al., 2019). We generated the input alignments for GEMME using the MMseqs2-based protocol implemented in ColabFold (Mirdita et al., 2022, Steinegger and Söding, 2017).
- ProteoCast classified the variants into impactful, uncertain or neutral by fitting a Gaussian Mixture Model to GEMME raw score distribution for each protein. Any residue with more than 10 (out of 19) impactful substitutions is considered as sensitive to mutations.
- ProteoCast applied the FPOP segmentation algorithm (Maidstone et al., 2017) on GEMME sensitivity profile (per-residue average scores), taking AlphaFold pLDDT into account. Segments that stand out from their surrounding background represent potential binding sites and regulatory motifs.
-
We validated of our proteome-wide predictions using different approaches.
First, by assessing the occurence of all missense mutations amongst two different resources for genetic variation in natural populations. The Drosophila melanogaster Genetic Reference Panel (Mackay et al., 2012) gives us access to natural polymorphism fixed in 200 inbred lines while the Drosophila Evolution over Space and Time (Kapun et al., 2021; Nunez et al. , 2024) gives us access to natural polymorphism occuring in the wild (sampling period 2009 - 2021). Second, by confronting our predictions to the FlyBase dataset of developmentally lethal mutations and referenced hypomorphs. Finally, we use existing databases for protein Post Translational Modifications and Short Linear Motifs to interpret residue mutational sensitivity.